You have access to two multiple sequence alignment programs in your lists: MAFFT and ClustalW. You can apply it to your lists. Note: If you have a list with many contigs, these programs may take along time to finish.
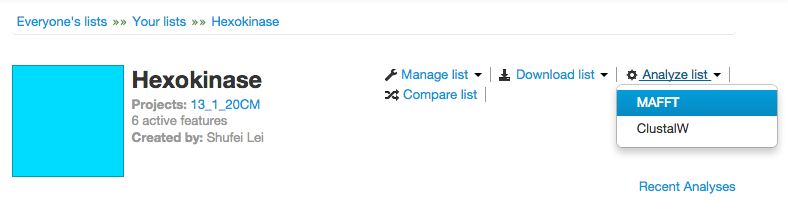
After you have selected a list, you will see the “Analyze list” dropdown menu.
MAFFT
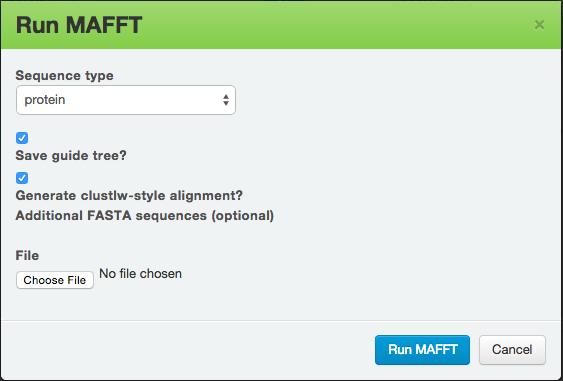
MAFFT is a multiple alignment program for amino acid or nucleotide sequences. After you have selected MAFFT, you will be presented with the following dialogue box:
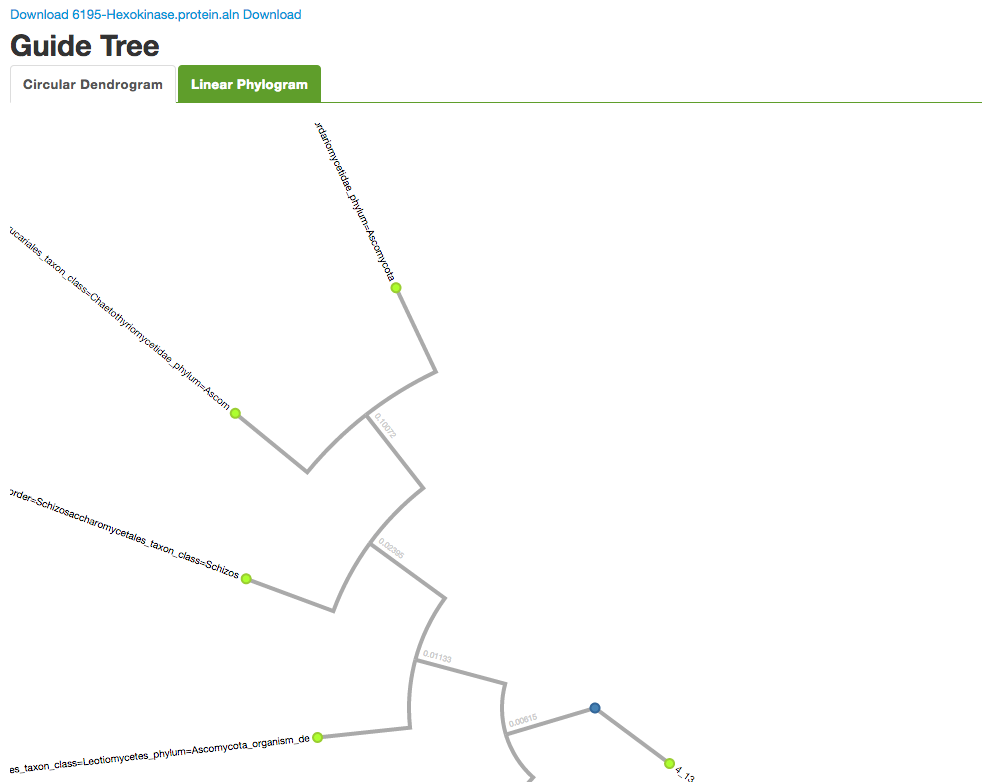
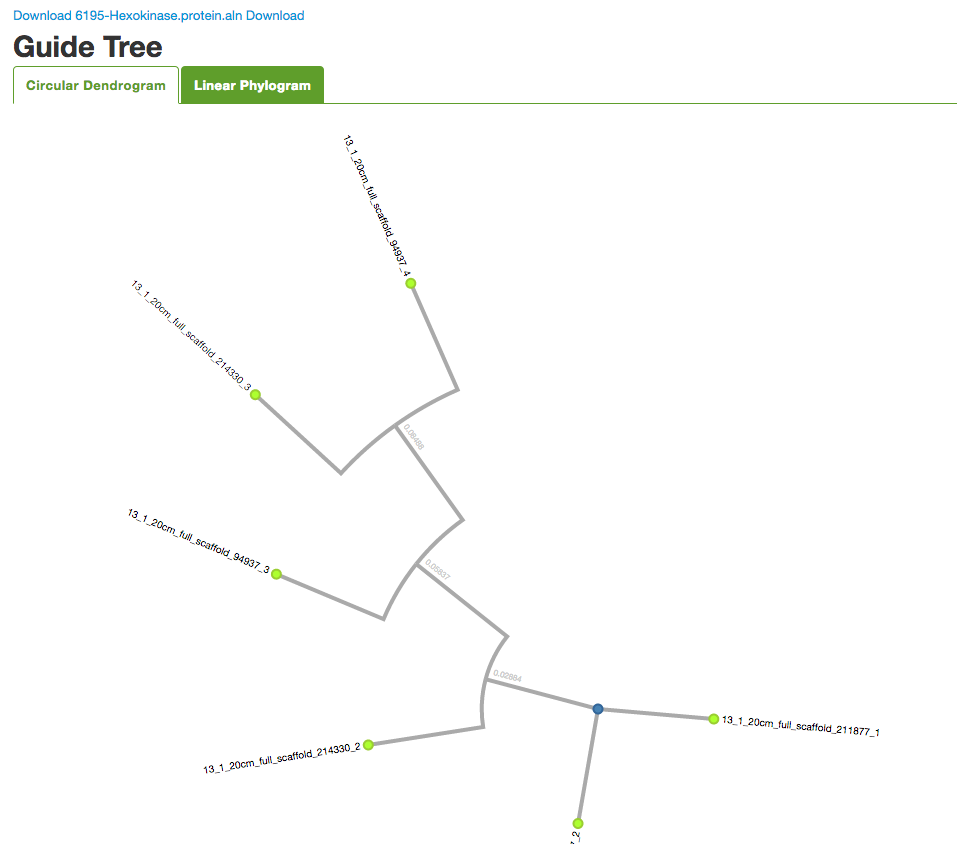
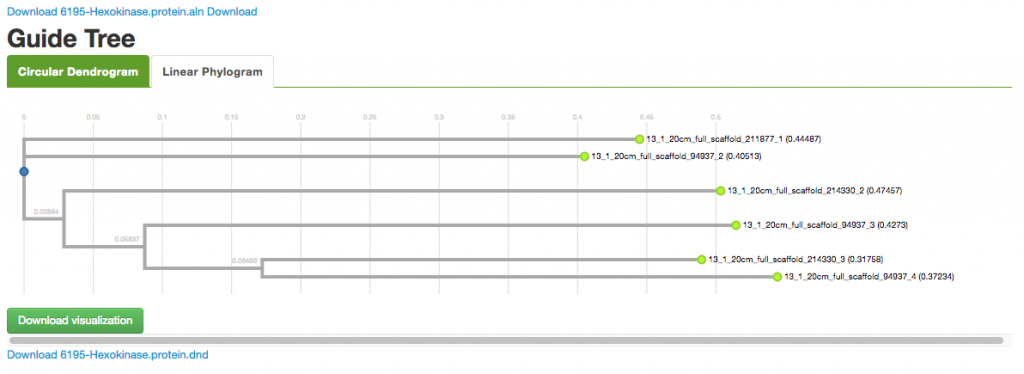
You can select Sequence type and check additional options. Click on Run MAFFT to start the program. The results provide you with the alignment sequences, a circular dendrogram, and a tree diagram.
You can use the links provided to download the protein sequences in .aln and .fasta formats, as well as the visualizations.
Clustalw
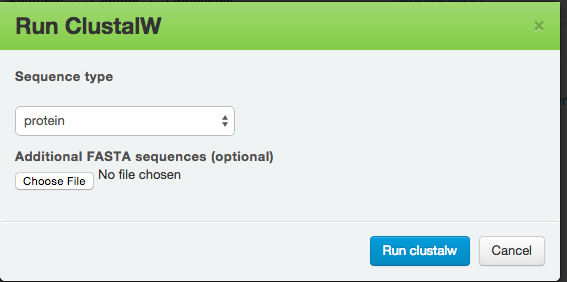
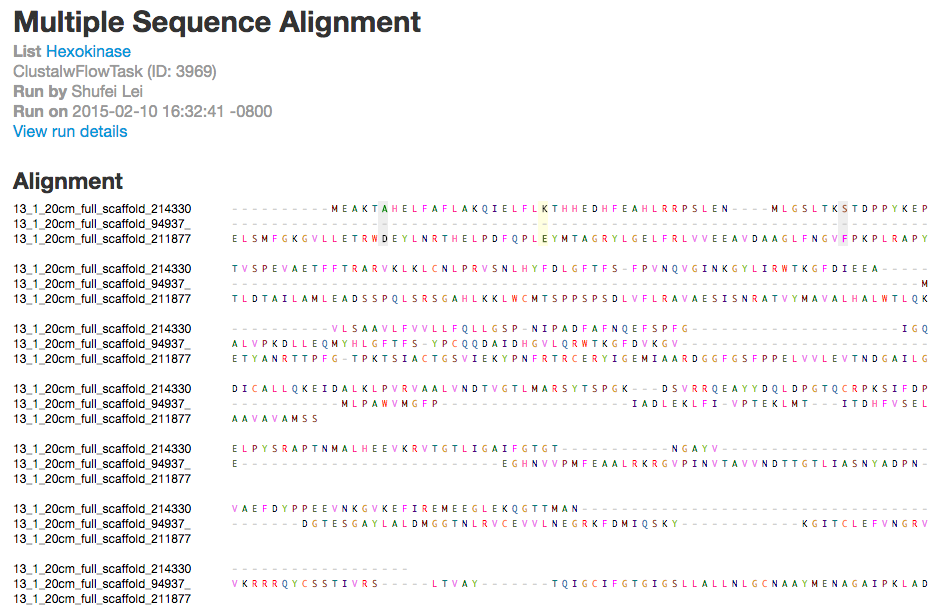
Similarly, ClustalW is a general purpose DNA or protein multiple sequence alignment program. After you have selected ClustalW, you will be presented with the following dialogue box:
And you also get the results in alignment sequences, circular dendrogram, and tree, along with all the download links.