You may want to download data and visualizations from ggKbase for the following purposes.
- Store data locally on your computer
- Analyze data using other analysis programs
- Share data with colleagues
There are a number of ways that you can download data from ggKbase.
**If you want to download data for many organisms or projects at once, instead of clicking through all those pages, slack Lily to do it on the backend**
How to Download
To trigger download, you need to first update the data file by clicking on the red exclamation mark, which indicates that the download needs to be generated. The symbol will turn into a yellow exclamation mark during the generation process. Once the file is generated/updated, the symbol will turn into a green check circle, which indicates that the file is now ready to download.
Places to Download Data File
Project Information Page
The central location for downloading ggKbase data in various data/file types is found in the project landing/home page, e.g. http://ggbkase.berkeley.edu/[project-slug]. Besides typing in the project link manually, if you are traversing through the project hierarchy, you will see the link at the breadcrumb:
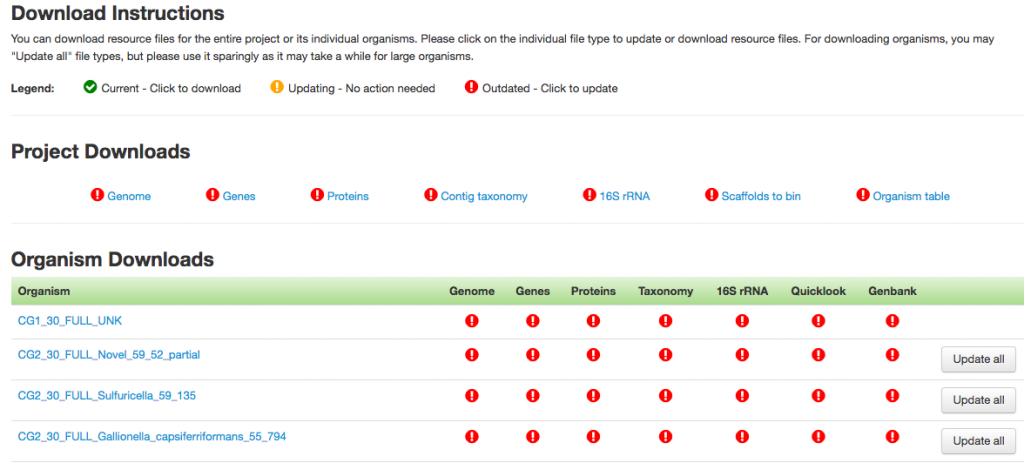
Below is a screenshot of the project page that shows you the instructions for downloading files. The download links are grouped by project level and organism level data files.
There are two other locations in which you can download data files.
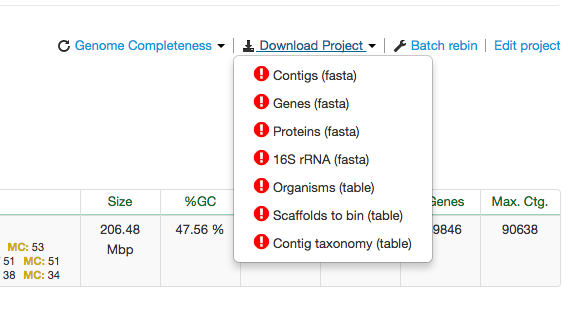
Organisms List Page
The Organisms list page provide you a dropdown menu to download the project level data files. Look for the Download dropdown menu like this:
When downloaded, these files are named as such:
Contigs(fasta) 14_0903_02_30cm.contigs.fa.gz Genes(fasta) 14_0903_02_30cm.genes.fna.gz Proteins(fasta) 14_0903_02_30cm.proteins.faa.gz 16S rRNA (fasta) 14_0903_02_30cm.16S.fna Organisms (table) 14_0903_02_30cm.organism_info.tsv Scaffolds to bin (table) 14_0903_02_30cm.scaffolds_to_bin.tsv Contig taxonomy (table) 14_0903_02_30cm.contig-taxonomy.tsv
Examples of the first few lines in each file:
Contigs(fasta) >14_0903_02_30cm_scaffold_22000 id=35997450 bin="14_0903_02_30cm_UNK" TCATGCAGCTGACGACAACGCACCCGGCGCGCTGCGCCCAGCGCATCGCCGCGCTCGACC TGGTCAGCAACGGGCGCGTCGAGTTCGCCACCGGCGAATCTGCCAGCATCACCAATTGAG >14_0903_02_30cm_scaffold_22010 id=35997460 bin="14_0903_02_30cm_UNK" GAGCCGGCGCCAGGCGCGTCAGCGCGGCCAGGGTCATTTCCGTTTCGCCGAAGCGCACCG GTCAGCGGCAGCTCGATGTCGAGAATCAGGTCCAGGTTCGCCGGCGTGTTGGCGGCAGCG Genes(fasta) >14_0903_02_30cm_scaffold_22000_1 Reverse transcriptase-RNase H-integrase n=1 Tax=Rhodotorula glutinis (strain ATCC 204091 / IIP 30 / MTCC 1151) RepID=G0SUI9_RHOG2 id=147996927 bin="14_0903_02_30cm_UNK" species=Rhodosporidium toruloides genus=Rhodosporidium taxon_order=Sporidiobolales taxon_class=Microbotryomycetes phylum=Basidiomycota organism_tax=unknown ATGGCTAAACCAATGAGACCCAGCGAATATGATGGAAAAACTCGTGACGCTCGAACTGTC GAAGCATGGCTTATTAGAATGACCACGTATTTGACGCTTACTAACACTGCGGACAATCGA >14_0903_02_30cm_scaffold_22010_4 Cobyrinic acid ac-diamide synthase; K04562 flagellar biosynthesis protein FlhG id=12556372 bin=CNBR_ACIDO species=Holophaga foetida genus=Holophaga taxon_order=Holophagales taxon_class=Holophagae phylum=Acidobacteria tax=CNBR_ACIDO organism_group=Acidobacteria organism_desc=why is coverage listed as 1? id=147997008 bin="14_0903_02_30cm_UNK" species=BJP_IG2102_Syntrophobacterales_60_12 genus=unknown taxon_order=Syntrophobacterales taxon_class=Deltaproteobacteria phylum=Proteobacteria organism_tax=unknown ATGAGTCCCACCCCCACGTCCCCGCGCCGCCCGATCAGCATCGCCGTCACGAGCGGCAAG GGGGGCGTTGGCAAGACCAGCGTCGCCGTGAACCTGGCGGTCGCGTTGGCGCGGCTGCGC Proteins(fasta) >14_0903_02_30cm_scaffold_22000_1 Reverse transcriptase-RNase H-integrase n=1 Tax=Rhodotorula glutinis (strain ATCC 204091 / IIP 30 / MTCC 1151) RepID=G0SUI9_RHOG2 id=147996927 bin="14_0903_02_30cm_UNK" species=Rhodosporidium toruloides genus=Rhodosporidium taxon_order=Sporidiobolales taxon_class=Microbotryomycetes phylum=Basidiomycota organism_tax=unknown MAKPMRPSEYDGKTRDARTVEAWLIRMTTYLTLTNTADNRKVELASSYLAGDAFEWYIDN QTVLLVGTFDGFKTALRDRFVPQNHKSITYSQYKGLTQGNLSISEYSIKFKALADQIPDL >14_0903_02_30cm_scaffold_22010_3 Tax=RIFCSPLOWO2_12_FULL_Acidobacteria_67_14b_curated id=147997007 bin="14_0903_02_30cm_UNK" species=RIFCSPLOWO2_12_FULL_Acidobacteria_67_14b_curated genus=unknown taxon_order=unknown taxon_class=unknown phylum=Acidobacteria organism_tax=unknown MRMTPREIDHTERDRLVVAHIGLVKALAHRLAQRLPPQVEIPDLISIGVLGLMDAASRYR ASLGVPFDAFARRRVQGAMLDALRELDWAPRSLRKLRREPTEEEIAAELNMTPAAYGRSL 16S rRNA (fasta) >14_0903_02_30cm_scaffold_381542_16S_1 16S ribosomal RNA (16S rRNA) id=149423351 bin="14_0903_02_30cm_UNK" organism_tax=unknown GAGAGACAGGTGCTGCATGGCTGTCGTCAGCTCGTGTCGTGAGAGGTTGGGTTAAGTCCC GCAACGAGCGCAACCCTTGCCTTTAGTTGCCATCATTCAGTTGGGCACTCTAAAGGGACT >14_0903_02_30cm_scaffold_383665_16S_1 16S ribosomal RNA (16S rRNA) id=149423352 bin="14_0903_02_30cm_UNK" organism_tax=unknown AGGCCCCTAAGGAGTGACTGGTGACTGGGGTGAAGTCGTAACAAGGTAGCCGTAGGGGAA AAGGCCCCTAAGGAGTGACTGGTGACTGGGGTGAAGTCGTAACAAGGTAGCCGTAGGGGA Organisms (table) name code taxonomy description bin length GC% coverage # contigs # features longest contig RP Inventory (total: 55) RP multiple BSCG Inventory (total: 51) BSCG multiple ASCG Inventory (total: 38) ASCG multiple curation status completion status 14_0903_02_30cm_UNK 14_0903_02_30cm_UNK unknown 832801812 62.53 5.11 458951 1171925 98379 55 55 51 51 35 32 Uncurated genome megabin 14_0903_02_30cm_Sphingomonadales_156_68_15 14_0903_02_30cm_Sphingomonadales_68_15 Sphingomonadales, Alphaproteobacteria, Proteobacteria, Bacteria 3028871 67.56 14.51 3 3046 2650155 52 1 51 0 12 0 Uncurated genome near complete Scaffolds to bin (table) scaffold_name bin organism taxonomy 14_0903_02_30cm_scaffold_22000 14_0903_02_30cm_UNK unknown 14_0903_02_30cm_scaffold_2884 14_0903_02_30cm_Solirubrobacterales_J_71_8 Actinobacteria, Actinobacteria, Bacteria Contig taxonomy (table) Contig name Size (bp) Coverage GC % Taxonomy winner Winner % Species winner Species winner % Genus winner Genus winner % Order winner Order winner % Class winner Class winner % Phylum winner Phylum winner % Domain winner Domain winner % 14_0903_02_30cm_scaffold_22000 5297 18.84 46.65 Rhodosporidium toruloides 1.0 Rhodosporidium toruloides 1.0 Rhodosporidium 1.0 Sporidiobolales 1.0 Microbotryomycetes 1.0 Basidiomycota 1.0 Fungi 1.0 14_0903_02_30cm_scaffold_22001 10189 7.2 68.81 Bacteria 1.0 RIFCSPLOWO2_12_FULL_RIF_CHLX_71_12_curated 0.33 unknown 0.75 unknown 0.83 unknown 0.75 Chloroflexi 0.33 Bacteria 1.0
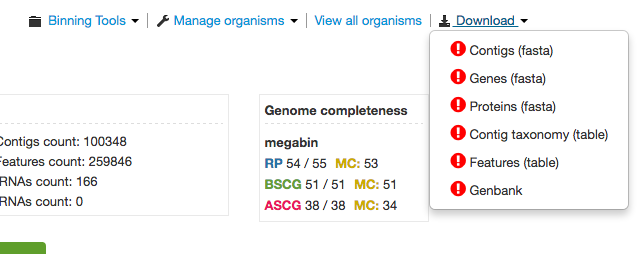
Individual Organism Page
Likewise, the individual Organism landing page also provides a dropdown list of download links but for an individual organism’s download files.
When downloaded, the file names are as such:
Contigs(fasta) 14_0903_02_30cm_Euryarchaeota_215_64_24.contigs.fa Genes(fasta) 14_0903_02_30cm_Euryarchaeota_215_64_24.genes.fna Proteins(fasta) 14_0903_02_30cm_Euryarchaeota_215_64_24.proteins.faa Contig taxonomy (table) 14_0903_02_30cm_Euryarchaeota_215_64_24.contig-taxonomy.tsv Features (table) 14_0903_02_30cm_Euryarchaeota_215_64_24.ql Genbank 14_0903_02_30cm_Euryarchaeota_215_64_24.gbk
Examples of the first few lines in each file:
Contigs(fasta)
>14_0903_02_30cm_scaffold_22369 id=35997819 bin="14_0903_02_30cm_Euryarchaeota_215_64_24"
CCCCTTATGTGAATGACTACGCCTTCCTCTGGGAGAGCGACCGAGCGACAGGGATTTCCG
CCGGGCTCCCGGCGGGATCGTGCCACGTCGTGCGCCATCCGTTCGACCCCTTTTTCCTCG
>14_0903_02_30cm_scaffold_4098 id=35998548 bin="14_0903_02_30cm_Euryarchaeota_215_64_24"
ATCCACGTGCTCGCTCACGTCGCCTTGGACACCGATCACGCCGATTCGCATCGGTCGACT
TAGGGCGGCAGCCGATTAAAACGATTTGGGGACGCCCGGTCGTTCAGCTGTTCTCGCGGT
Genes(fasta)
>14_0903_02_30cm_scaffold_22369_1 hypothetical protein n=1 Tax=Rhodocyclaceae bacterium RZ94 RepID=UPI00037C713E id=148003626 bin="14_0903_02_30cm_Euryarchaeota_215_64_24" organism_tax=Euryarchaeota, Archaea
CCTTATGTGAATGACTACGCCTTCCTCTGGGAGAGCGACCGAGCGACAGGGATTTCCGCC
GGGCTCCCGGCGGGATCGTGCCACGTCGTGCGCCATCCGTTCGACCCCTTTTTCCTCGAT
>14_0903_02_30cm_scaffold_22369_2 hypothetical protein id=148003627 bin="14_0903_02_30cm_Euryarchaeota_215_64_24" organism_tax=Euryarchaeota, Archaea
ATGGTTCCCTCCGAACGACCTCCCGCGGCCGGTGCGACCTTCCCGTACATCGGCCTCGCG
GTGGCCGTCCTCGCACTGTATGCGATCCTCGCGGTCACGATGCCTCTGAATCCCTATCGG
Proteins(fasta)
>14_0903_02_30cm_scaffold_22369_1 hypothetical protein n=1 Tax=Rhodocyclaceae bacterium RZ94 RepID=UPI00037C713E id=148003626 bin="14_0903_02_30cm_Euryarchaeota_215_64_24" organism_tax=Euryarchaeota, Archaea
PYVNDYAFLWESDRATGISAGLPAGSCHVVRHPFDPFFLDRRGAGLGSRLSGAFGPVPRR
VIFSGPPEATRGSRDVVQLPSVLPRDPPTQVVLLLRDGRFPAPVVKRKRIGVHEVLAVHG
>14_0903_02_30cm_scaffold_22369_2 hypothetical protein id=148003627 bin="14_0903_02_30cm_Euryarchaeota_215_64_24" organism_tax=Euryarchaeota, Archaea
MVPSERPPAAGATFPYIGLAVAVLALYAILAVTMPLNPYRAAVALVAFFAMGYCTLGLVA
GGRIPMSVAEILAFTVGLTILITALSALAVSIVGIPITEFAVVIVGLPLAVIAFLLRRPA
Contig taxonomy (table)
Contig name Size (bp) Coverage GC % Taxonomy winner Winner % Species winner Species winner % Genus winner Genus winner % Order winner Order winner % Class winner Class winner % Phylum winner Phylum winner % Domain winner Domain winner %
14_0903_02_30cm_scaffold_4098 12145 17.44 60.35 Archaea 0.91 RBG_19FT_COMBO_Euryarchaeota_69_17_curated 0.27 unknown 0.55 unknown 0.55 unknown 0.55 Euryarchaeota 0.45 Archaea 0.91
14_0903_02_30cm_scaffold_4370 11798 30.33 64.66 Euryarchaeota 0.94 RBG_19FT_COMBO_Euryarchaeota_69_17_curated 0.47 unknown 1.0 unknown 1.0 unknown 1.0 Euryarchaeota 0.94 Archaea 0.94
Features (table)
14_0903_02_30cm_scaffold_22369_1 14_0903_02_30cm_scaffold_22369 1 5248 63.99 21.49 11 3 704 u 5.2e-09 69 hypothetical protein n=1 Tax=Rhodocyclaceae bacterium RZ94 RepID=UPI00037C713E unknown smd:Smed_5686;UniRef100_UPI00037C713E
14_0903_02_30cm_scaffold_22369_4 14_0903_02_30cm_scaffold_22369 4 5248 63.99 21.49 11 3662 4219 c 6.6e-37 162 Tax=RBG_16_Euryarchaeota_68_13_curated RBG_16_Euryarchaeota_68_13_curated, unknown, unknown, unknown, Euryarchaeota, Archaea 86939202
Genbank
LOCUS 14_0903_02_30cm_scaffold_22369 5248 bp DNA linear BCT MAR-29-2017
DEFINITION 14_0903_02_30cm_scaffold_22369, contig.
ACCESSION 14_0903_02_30cm_scaffold_22369
VERSION 14_0903_02_30cm_scaffold_22369.1 GI:
KEYWORDS .
SOURCE 14_0903_02_30cm_Euryarchaeota_215_64_24
ORGANISM 14_0903_02_30cm_Euryarchaeota_215_64_24
COMMENT Data sourced from ggkbase. For additional details see
http://ggkbase.berkeley.edu/organisms/47637
FEATURES Location/Qualifiers
source 1..5248
/organism="14_0903_02_30cm_Euryarchaeota_215_64_24"
/mol_type="genomic DNA"
gene <3..704
/locus_tag="14_0903_02_30cm_scaffold_22369_1"
CDS <3..704
/product="hypothetical protein n=1 Tax=Rhodocyclaceae
bacterium RZ94 RepID=UPI00037C713E"
/codon_start=3
/transl_table=11
/translation="PYVNDYAFLWESDRATGISAGLPAGSCHVVRHPFDPFFLDRRGA
GLGSRLSGAFGPVPRRVIFSGPPEATRGSRDVVQLPSVLPRDPPTQVVLLLRDGRFPA
PVVKRKRIGVHEVLAVHGLITREELRAIYRTSHVAVFPYRFVRTGLPLVVLEAVAAGL
PVVTTRIHPIRELEGRTGLVFARPRDPPDIARAIESAFDDAQRAAVVRKNDEWIRTTP
DWSTVAKNFVSFVRR*"
/db_xref="smd:Smed_5686"
/db_xref="UniRef100_UPI00037C713E"
gene 770..1669
/locus_tag="14_0903_02_30cm_scaffold_22369_2"
CDS 770..1669
/product="hypothetical protein"
/codon_start=1
/transl_table=11
/translation="MVPSERPPAAGATFPYIGLAVAVLALYAILAVTMPLNPYRAAVA
LVAFFAMGYCTLGLVAGGRI
/db_xref="86935684" gene complement(4247..4693)
/locus_tag="14_0903_02_30cm_scaffold_22369_5"
CDS complement(4247..4693)
/product="Tax=RBG_16_Euryarchaeota_68_12_curated"
/codon_start=1
/transl_table=11
/translation="MGRESSDALEQAALSFLSGIDPDLGLDAALFVRRTGLVLASWMR
EGIRLDVVSVMAATMLASVDTIIESVGGPTPEVISVDTDAHQILATKVNSRAFLVVIA
PKKVSRTVVRKTMRGLNARLAAAASKSTHLHVEETEKQRVNVRPPR*"
/db_xref="86935684"
ORIGIN
1 CCCCTTATGT GAATGACTAC GCCTTCCTCT GGGAGAGCGA CCGAGCGACA GGGATTTCCG
61 CCGGGCTCCC GGCGGGATCG TGCCACGTCG TGCGCCATCC GTTCGACCCC TTTTTCCTCG
121 ATCGCAGGGG CGCGGGCCTC GGATCCCGAC TGTCCGGGGC CTTCGGACCC GTTCCTCGGC
181 GCGTCATCTT CTCGGGGCCT CCCGAGGCCA CTCGGGGGAG CCGCGACGTG GTCCAGCTTC
241 CGAGCGTCCT TCCCCGGGAC CCTCCGACGC AGGTCGTCCT GCTCCTGCGT GACGGGCGAT
301 TTCCCGCCCC GGTCGTCAAG CGGAAGCGGA TCGGCGTCCA CGAGGTCCTC GCCGTCCACG
361 GCCTGATCAC GCGGGAGGAG TTGCGCGCGA TCTACCGCAC ATCCC
Note- the Features Table for a given Organism contains:
feature_ID, contig_ID, feature_number, contig_length, GC%, coverage, codon table, winning_taxonomy_level, begin_position, end_position, complement (uncomplemented or complemented), E-value, bit_score, value_of_annotation
In order to download the 16S for a specific organism for example, click on “rRNA” for that organism, then click on the feature which may then be downloaded.
Why are some files *.fa and others *.fna or *.fasta?
On biotite and ggKbase:
.fa or .fasta – contig or genome sequences
.fna – gene sequences