ggKbase provides a BLAST (Basic Local Alignment Search Tool) program for sequence comparison. However, this tool is only accessible after you have signed into your account.
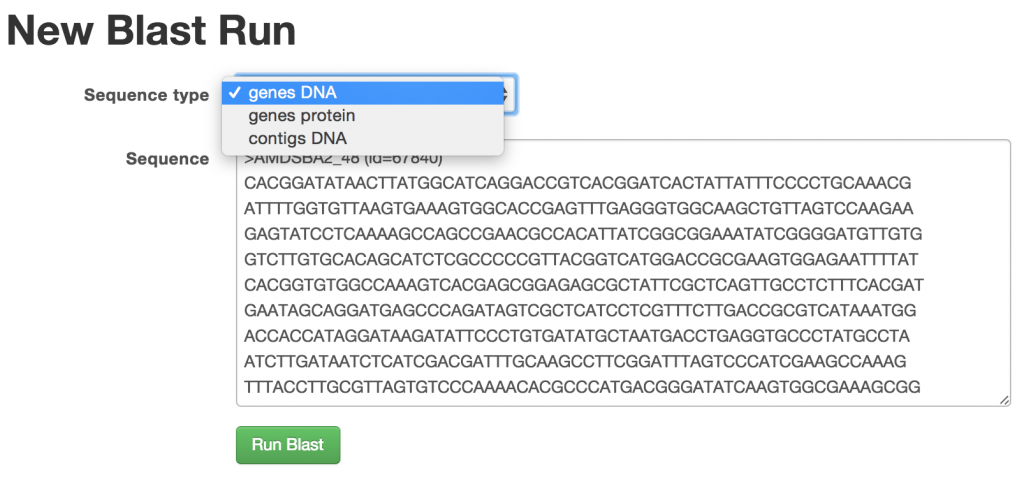
BLAST Form
The input sequence type can be genes DNA, genes protein, or contigs DNA.
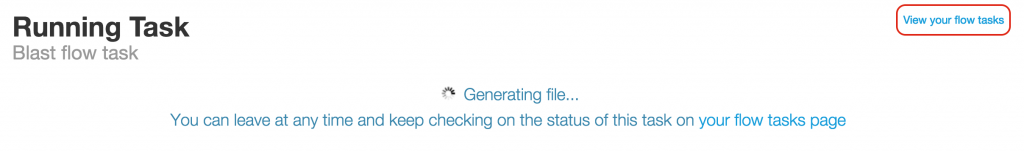
BLAST Flow Task
Once you click on the green “Run Blast” button, you will be shown the Blast flow task processing page with the message “Generating file…” This page refreshes itself until the task is done. This process may take a few minutes; however, you don’t need to wait on this page until the task is done. You can check on the “View your flow tasks” link to check on the status of your BLAST flow task.
And if you have moved on to other pages on the site, you can always click on the dropdown menu on the upper right hand corner and then click on “My flow tasks.”
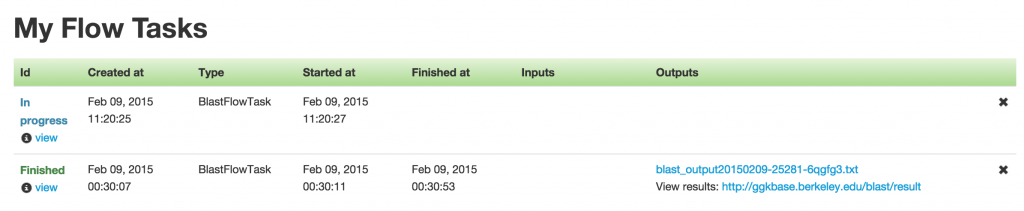
The BLAST Flow Task page (see below) shows a history of past finished tasks as well as those that are being processed at the moment. The finished tasks contain links to the BLAST results.
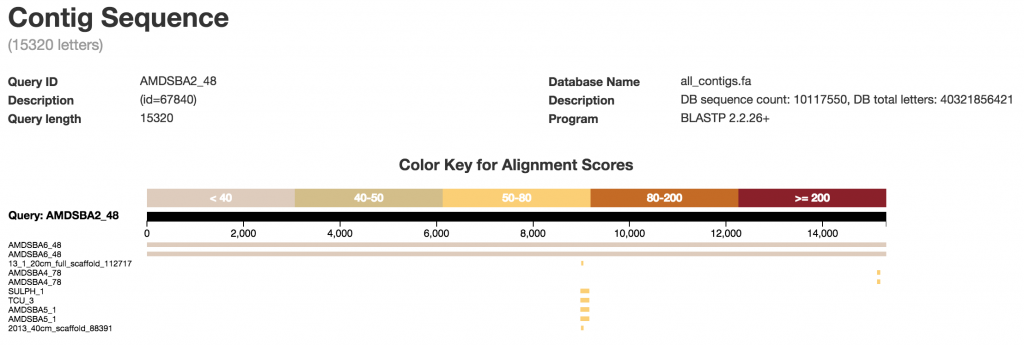
BLAST Results
The BLAST results are displayed in visualization of the alignment scores.
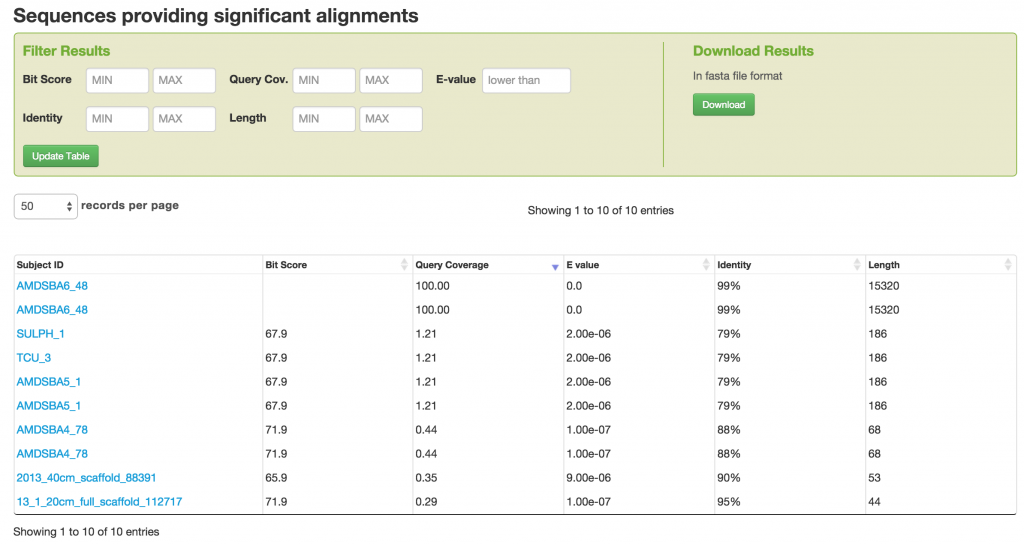
We can interact with the BLAST results by filtering them. You can also download the Blast results in FASTA file format.
Alternative Way to use BLAST
Alternatively, you can Blast a sequence by going to their individual contig or gene page.
BLAST at the Contig Page
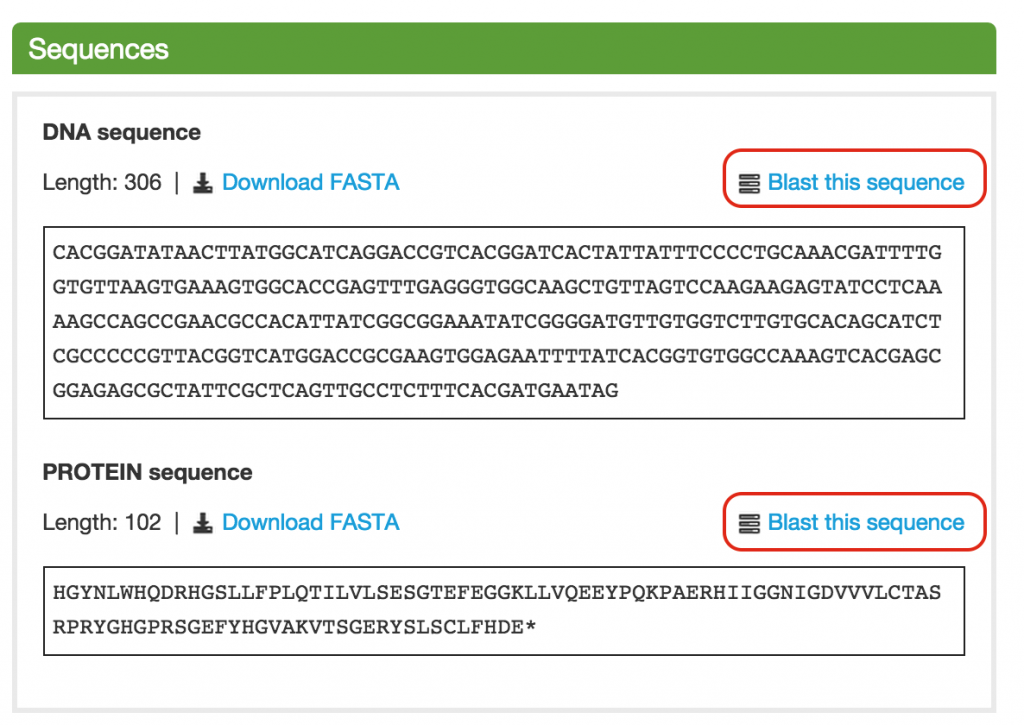
On the upper right hand side, you can find the “Blast sequence” link. This link will take you to the BLAST page with the sequence box filled in with the contig’s sequence.
BLAST at the Gene/Feature Page
In the Sequences section on the Gene page, you can find the “Blast this sequence” link. This link will take you to the BLAST page with the sequence box filled in with the DNA or Protein sequence of the gene.

One thought on “BLAST”
Comments are closed.