One of the primary analysis tools in ggKbase is the Lists tool. Lists are collections of features / genes that match a set of search terms, which often try to capture entire metabolic pathways, e.g. Glycolysis. Lists are dynamic because they change when data in ggKbase is updated, e.g. new set of annotations, user-created notes. Lists contents can be visualized in the Genome Summary page.
You can browse everyone’s lists by click on the Lists link on the top menu.
You can also view only your lists by clicking on the link to “Your lists” on the right at the same level as the page title.
This section focuses on how to create a new list. So, click on the link “Create a new list” to access the “create a new list” form.
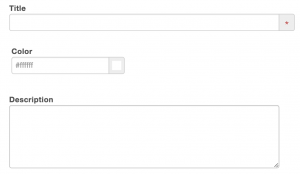
Step 1: Title, Color, Description
As a starter, give a title to the list that you are about create. You may set a color for the list. You may also provide more context and information about the list in the description box.
Step 2: Hide, Lock
Depending on whether you want to show your list to everyone in ggKbase, you may choose to hide it by clicking on the “Hidden” checkbox.
And you may protect your lists by accidental tampering, you may lock your list by clicking on the “Locked” checkbox. Note: Even the list owner cannot change the list once it’s locked.
Step 3: Search by Keywords
The main mechanism for finding the desired collection of genes is to use the search boxes.
The three search boxes use boolean logic. The first box produces genes that have annotations matching one or more of the keywords (boolean OR). The second search box requires the genes annotation to match all the keywords (boolean AND). The last search box allows you to enter undesired keywords, excluding genes that have these keywords in their annotations (boolean NOT).
Step 4: Search Scope by Projects
You can also set the scope of your search by projects.
Step 5: Save List
The last step is to click on the blue “Save List” button on the bottom of the page.
By saving the list, you can modify it in the future. Also, you can use it in the Genome Summary page.
Alternative Way to Create a List
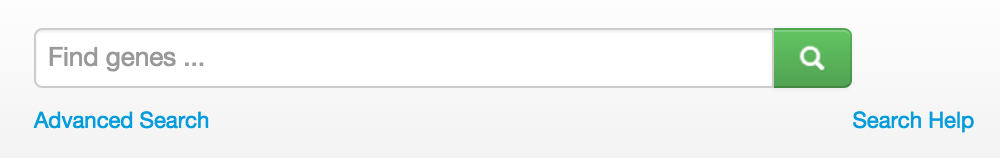
Another way to create a list is to start with the main search box on the top banner. It searches among the annotations, notes and systematic names (DB x-refs); therefore, you won’t find features by name using this search box. For more information about the search functions, click here.
After you click on the search button, you will be taken to the “Advanced Feature Search” page where you can set the scope of the search by projects, similar to the directions above.





2 thoughts on “Lists”
Comments are closed.